DTI Analysis Techniques
Many different DTI analysis techniques and approaches have been applied to study a range of pathologies and include region of interest analysis, tractography, histogram analysis, atlas-based segmentation, quantification of graph-based connectivity networks, and voxel-based analysis to name but a few. Each of these techniques has its own strengths and limitations and there is no single technique that can be regarded as superior to all the others. The most optimal analysis approach depends on many factors, including:
- The purpose of the analysis (e.g., to delineate a known fibre bundle, to explore the data)
- Whether it is for a single subject or group comparison
- The data quality
- …
- Whole-brain analyses
- Region-specific analyses
- Voxel-based analyses
Whole-Brain Analysis Techniques
The general concept of whole brain DTI analysis techniques is to obtain quantitative DTI measures from all the voxels that include brain white matter, and can thus be subdivided into two parts:- An approach to define which voxels are part of the brain white matter
- An approach to extract relevant DTI information from these voxels
Histogram Analysis
Once the DTI measures have been extracted from the selected voxels of interest, they can be summarized using a histogram. This histogram is a frequency distribution that displays the number of voxels with a specific value of the diffusion measure (e.g., FA). Whole-brain analysis of DTI data has the following strengths and limitations:
- Less reliant on user intervention than other approaches
- Results obtained quickly, without labor-intensive interventions
- Regional information is lost as DTI measures are averaged over the whole-brain white matter
- Results are sensitive to partial volume effects due to atrophy
- Results can depend on segmentation/registration accuracy or whole-brain tractography parameters
Region-Specific Analysis Techniques
In region specific analysis techniques, diffusion measures are obtained in one or more predefined areas of the brain. DTI measures, such as the FA and MD are thus statistically evaluated in an anatomical region or white matter tract reconstruction. There are two main approaches:
- Region of interest analysis
- Tractography analysis
Region Of Interest Analysis
In region of interest (ROI) analysis , diffusion measures are obtained from a specific brain region, which is defined by manual delineation or by automated segmentation or parcellation. As automated segmentations are less observer dependent and thus more reproducible, they have some clear advantages over manual delineations. However, automated segmentations are not always appropriate, for example due to ill-defined boundaries in regions of pathology. Instead of delineating regions and structures manually, automatic segmentation methods can be used. Such automated methods are especially useful when structures or lesions can be accurately segmented on the anatomical MRI. As an example, T2 lesions could be segmented in a patient with multiple sclerosis. After registering the T1/T2 MR image to the DTI data set and applying the deformation field to the segmented lesion masks, DTI measures can be derived from these lesions. Bear in mind that these results will strongly depend on the segmentation and registration accuracy, especially when some of the lesions are small. In addition, the resolution of the DTI image is typicall y lower than the resolution of the anatomical MRI that is used for the segmentation, leading to partial volume effects.Region-specific analysis of DTI data by using the ROI approach has the following advantages and limitations:
- In comparison to whole-brain analyses, more regionally specific information is obtained
- Manual delineation is closer to the original data than other techniques which require more complex modeling and image processing
- ROI analysis is less dependent on parameter settings than tractography or voxel-based analysis
- Requires a prior hypothesis about where differences could be found, as that is where the ROI will be placed.
- Intra- and inter-observer reproducibility of results should be assessed, as manual delineation is subjective.
- Requires clear guidelines that describe how the ROI should be defined.
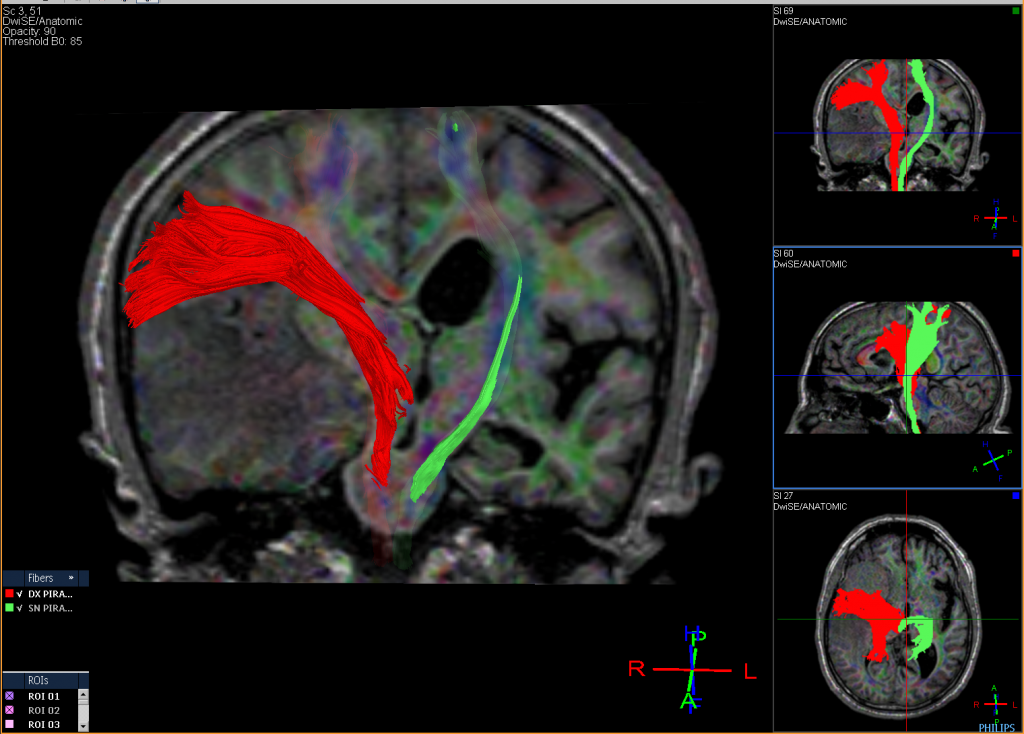
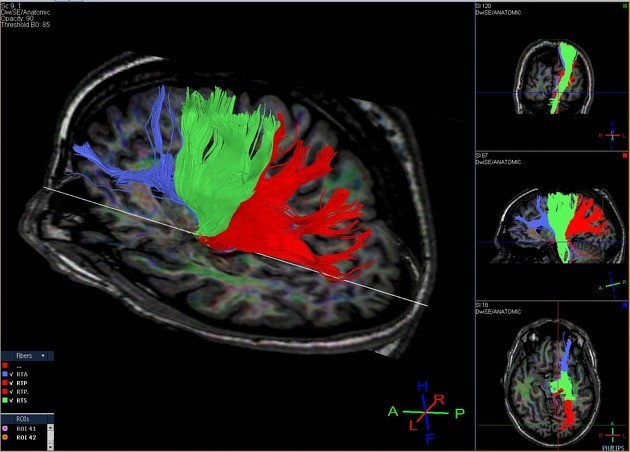
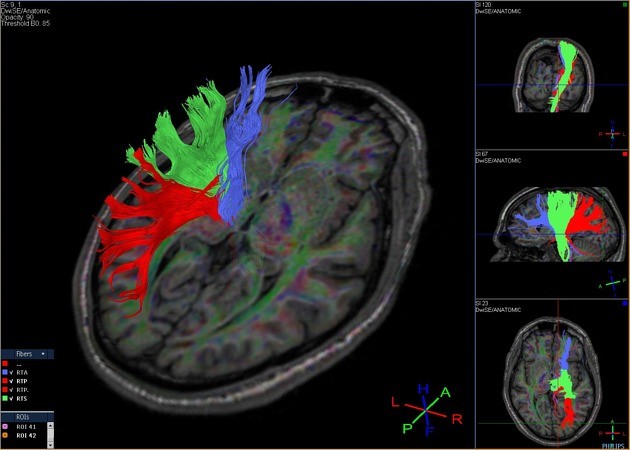
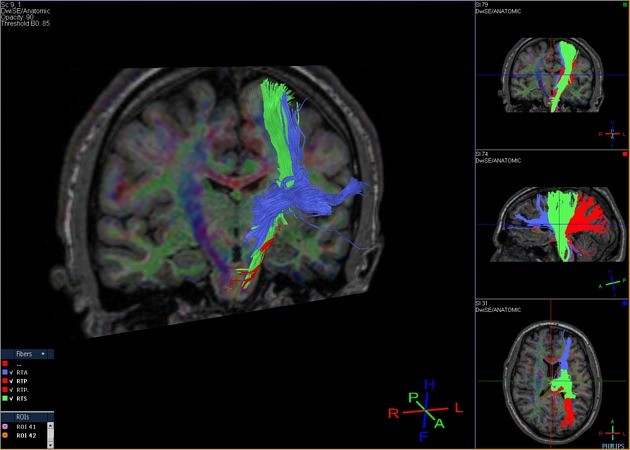
Tractography Analysis
The delineation of white matter tracts using only 2D manually drawn ROIs or anatomical MR images is not optimal for the reasons outlined previously. However, by using the inherent directional diffusion information in the DTI data set, virtual representations of white matter fibre bundles can be reconstructed, using tractography.
Diffusion tensor imaging (DTI) is a magnetic resonance imaging technique that enables the measurement of the restricted diffusion of water in tissue in order to produce neural tract images instead of using this data solely for the purpose of assigning contrast or colors to pixels in a cross-sectional image.
DTI provides information about structural connectivity, as compared to functional connectivity data from rsFMRI. Perhaps most importantly, DTI can illustrate to the surgeon the relationship of a tumor to underlying white matter tracts.
Diffusion tensor imaging tractography, or DTI tractography, is an MRI (magnetic resonance imaging) technique that measures the rate of water diffusion between cells to understand and create a map of the body’s internal structures; it is most commonly used to provide imaging of the brain.